一步筛选合成蛋白质
来源:《PNAS》
作者:Kelly M. Chacón等
时间:2014-03-12
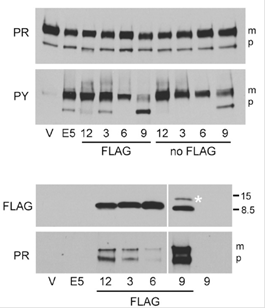
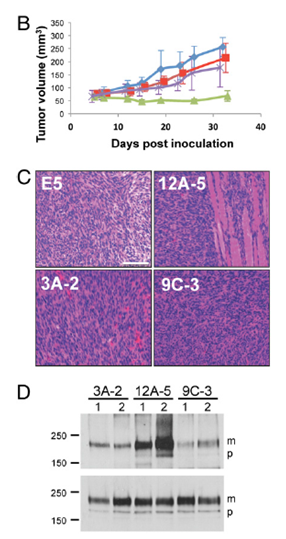
由病毒癌基因和缺乏天然存在的氨基酸序列的人工致癌基因产物可以形成肿瘤。
蛋白质通过突变和自然选择两个迭代过程实现进化,但可以以这种方式获得的蛋白质多样性则受开始原料约束并依赖于中间体的逐步选择。为了获得来自已有序列以外的、所需功能的蛋白质,耶鲁大学医学院Kelly Chacón等人设计了一种用于合成蛋白致癌活性的单步筛选方法。他们用简并寡核苷酸合成编码氨基酸随机链的DNA,以及克隆DNA为逆转录病毒表达文库,产生成百上千称为traptamers的 29氨基酸跨膜蛋白。感染文库的小鼠细胞产生少量转化位点。Kelly Chacón等恢复四个能够引起转化的traptamers,没有一个与任何已知的蛋白质同源。活小鼠细胞表达这些traptamers形成肿瘤,三个traptamers能够转化人成纤维细胞。四个traptamers都与血小板源性生长因子β受体有关,通过受体的配体独立激活诱导转化。这种称为从头选择的蛋白质工程可以应用于缺乏结构-功能详细信息或复杂模型的情况,有助于调节多样化生物通路。(编译:中国科学院成都生物研究所 王芋华,王海燕)
De novo selection of oncogenes
Abstract All cellular proteins are derived from preexisting ones by natural selection. Because of the random nature of this process, many potentially useful protein structures never arose or were discarded during evolution. Here, we used a single round of genetic selection in mouse cells to isolate chemically simple, biologically active transmembrane proteins that do not contain any amino acid sequences from preexisting proteins. We screened a retroviral library expressing hundreds of thousands of proteins consisting of hydrophobic amino acids in random order to isolate four 29-aa proteins that induced focus formation in mouse and human fibroblasts and tumors in mice. These proteins share no amino acid sequences with known cellular or viral proteins, and the simplest of them contains only seven different amino acids. They transformed cells by forming a stable complex with the platelet-derived growth factor β receptor transmembrane domain and causing ligand-independent receptor activation. We term this approach de novo selection and suggest that it can be used to generate structures and activities not observed in nature, create prototypes for novel research reagents and therapeutics, and provide insight into cell biology, transmembrane protein–protein interactions, and possibly virus evolution and the origin of life.
Significance Artificial proteins may have improved properties compared with proteins that arose during evolution, but approaches to construct active artificial proteins are cumbersome and often constrained by existing protein structures. Here, we used mouse cells to select proteins that formed tumors from a library of small transmembrane proteins with randomized hydrophobic amino acid sequences. The resulting oncoproteins lack amino acid sequences from any known protein and function by activating a cellular growth factor receptor. This approach can be used to generate structures not observed in nature, create prototypes for research and possibly clinical uses, and provide insight into cell biology, protein–protein interactions, and evolution.
原文链接:http://www.pnas.org/content/early/2013/12/12/1315298111.full.pdf+html




