疟原虫青蒿素抗性的分子标记
来源:《Nature》
作者:Frédéric Ariey等
时间:2014-03-12
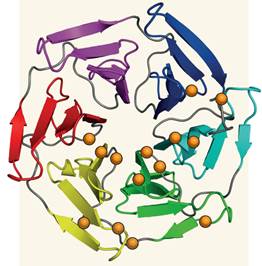
一系列关于离体、基因组、生态和流行病学的研究已经确定了恶性疟原虫在对抗青蒿素类抗疟药物时发挥关键作用的基因突变。
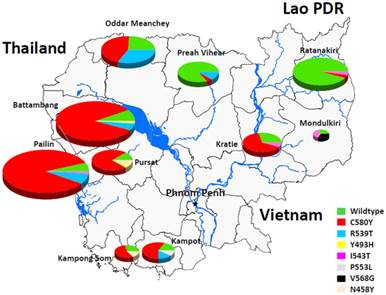
分子标记有助于监测东南亚青蒿素耐药性恶性疟原寄生虫。来自法国巴斯德研究所等研究机构的科学家们发现,无论是立体还是活体内,寄生虫基因组编码K13蛋白的propeller区域的突变与青蒿素耐药相关,也与柬埔寨各省流行的疟原虫抗性相关。(编译:中国科学院成都生物研究所 王芋华,王海燕)
A molecular marker of artemisinin-resistant Plasmodium falciparum malaria
Abstract Plasmodium falciparum resistance to artemisinin derivatives in southeast Asia threatens malaria control and elimination activities worldwide. To monitor the spread of artemisinin resistance, a molecular marker is urgently needed. Here, using whole-genome sequencing of an artemisinin-resistant parasite line from Africa and clinical parasite isolates from Cambodia, we associate mutations in the PF3D7_1343700 kelch propeller domain (‘K13-propeller’) with artemisinin resistance in vitro and in vivo. Mutant K13-propeller alleles cluster in Cambodian provinces where resistance is prevalent, and the increasing frequency of a dominant mutant K13-propeller allele correlates with the recent spread of resistance in western Cambodia. Strong correlations between the presence of a mutant allele, in vitro parasite survival rates and in vivo parasite clearance rates indicate that K13-propeller mutations are important determinants of artemisinin resistance. K13-propeller polymorphism constitutes a useful molecular marker for large-scale surveillance efforts to contain artemisinin resistance in the Greater Mekong Subregion and prevent its global spread.
原文链接:http://www.nature.com/nature/journal/v505/n7481/pdf/nature12876.pdf
下面是系列相关文献:
MALARIA Did They Really Say ... Eradication?
Summary SEATTLE, WASHINGTON-- The malaria world is all abuzz about a call by Bill and Melinda Gates to wipe the scourge from the planet. Even if it proves unfeasible, their idea could have a big impact.
原文链接:http://www.sciencemag.org/content/318/5856/1544.full.pdf
A Major Genome Region Underlying Artemisinin Resistance in Malaria
Abstract Evolving resistance to artemisinin-based compounds threatens to derail attempts to control malaria. Resistance has been confirmed in western Cambodia and has recently emerged in western Thailand, but is absent from neighboring Laos. Artemisinin resistance results in reduced parasite clearance rates (CRs) after treatment. We used a two-phase strategy to identify genome region(s) underlying this ongoing selective event. Geographical differentiation and haplotype structure at 6969 polymorphic single-nucleotide polymorphisms (SNPs) in 91 parasites from Cambodia, Thailand, and Laos identified 33 genome regions under strong selection. We screened SNPs and microsatellites within these regions in 715 parasites from Thailand, identifying a selective sweep on chromosome 13 that shows strong association (P = 10−6 to 10−12) with slow CRs, illustrating the efficacy of targeted association for identifying the genetic basis of adaptive traits.
原文链接:http://www.sciencemag.org/content/336/6077/79.full.pdf
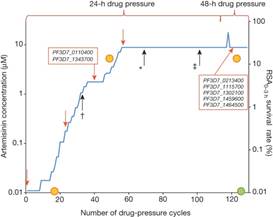
Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies
Summary Background Artemisinin resistance in Plasmodium falciparum lengthens parasite clearance half-life during artemisinin monotherapy or artemisinin-based combination therapy. Absence of in-vitro and ex-vivo correlates of artemisinin resistance hinders study of this phenotype. We aimed to assess whether an in-vitro ring-stage survival assay (RSA) can identify culture-adapted P falciparum isolates from patients with slow-clearing or fast-clearing infections, to investigate the stage-dependent susceptibility of parasites to dihydroartemisinin in the in-vitro RSA, and to assess whether an ex-vivo RSA can identify artemisinin-resistant P falciparum infections. Methods We culture-adapted parasites from patients with long and short parasite clearance half-lives from a study done in Pursat, Cambodia, in 2010 (registered with ClinicalTrials.gov, number NCT00341003) and used novel in-vitro survival assays to explore the stage-dependent susceptibility of slow-clearing and fast-clearing parasites to dihydroartemisinin. In 2012, we implemented the RSA in prospective parasite clearance studies in Pursat, Preah Vihear, and Ratanakiri, Cambodia (NCT01736319), to measure the ex-vivo responses of parasites from patients with malaria. Continuous variables were compared with the Mann-Whitney U test. Correlations were analysed with the Spearman correlation test. Findings In-vitro survival rates of culture-adapted parasites from 13 slow-clearing and 13 fast-clearing infections differed significantly when assays were done on 0–3 h ring-stage parasites (10·88% vs 0·23%; p=0·007). Ex-vivo survival rates significantly correlated with in-vivo parasite clearance half-lives (n=30, r=0·74, 95% CI 0·50–0·87; p<0·0001). Interpretation The in-vitro RSA of 0–3 h ring-stage parasites provides a platform for the molecular characterisation of artemisinin resistance. The ex-vivo RSA can be easily implemented where surveillance for artemisinin resistance is needed. Funding Institut Pasteur du Cambodge and the Intramural Research Program, NIAID, NIH.
原文链接:http://www.sciencedirect.com/science/article/pii/S1473309913702524#
Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia
Abstract The recent emergence of artemisinin-resistant Plasmodium falciparum malaria in western Cambodia could threaten prospects for malaria elimination. Identification of the genetic basis of resistance would provide tools for molecular surveillance, aiding efforts to contain resistance. Clinical trials of artesunate efficacy were conducted in Bangladesh, in northwestern Thailand near the Myanmar border, and at two sites in western Cambodia. Parasites collected from trial participants were genotyped at 8,079 single nucleotide polymorphisms (SNPs) using a P. falciparum-specific SNP array. Parasite genotypes were examined for signatures of recent positive selection and association with parasite clearance phenotypes to identify regions of the genome associated with artemisinin resistance. Four SNPs on chromosomes 10 (one), 13 (two), and 14 (one) were significantly associated with delayed parasite clearance. The two SNPs on chromosome 13 are in a region of the genome that appears to be under strong recent positive selection in Cambodia. The SNPs on chromosomes 10 and 13 lie in or near genes involved in postreplication repair, a DNA damage-tolerance pathway. Replication and validation studies are needed to refine the location of loci responsible for artemisinin resistance and to understand the mechanism behind it; however, two SNPs on chromosomes 10 and 13 may be useful markers of delayed parasite clearance in surveillance for artemisinin resistance in Southeast Asia.
原文链接:http://www.pnas.org/content/110/1/240.full.pdf+html
Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia
Abstract We describe an analysis of genome variation in 825 P. falciparum samples from Asia and Africa that identifies an unusual pattern of parasite population structure at the epicenter of artemisinin resistance in western Cambodia. Within this relatively small geographic area, we have discovered several distinct but apparently sympatric parasite subpopulations with extremely high levels of genetic differentiation. Of particular interest are three subpopulations, all associated with clinical resistance to artemisinin, which have skewed allele frequency spectra and high levels of haplotype homozygosity, indicative of founder effects and recent population expansion. We provide a catalog of SNPs that show high levels of differentiation in the artemisinin-resistant subpopulations, including codon variants in transporter proteins and DNA mismatch repair proteins. These data provide a population-level genetic framework for investigating the biological origins of artemisinin resistance and for defining molecular markers to assist in its elimination.
原文链接:http://www.nature.com/ng/journal/v45/n6/full/ng.2624.html




